Large File Formats
Ensembl supports a number of large (multi-gigabyte) file formats for datasets that are too large to upload directly to the Ensembl servers. The file remains on the remote server and is accessed via its URL, with only small chunks of data being requested by the Ensembl webcode at any one time.
Supported formats currently include:
All of these formats can be viewed in Ensembl by creating a custom data track using the 'Custom tracks' function, or by using a track hub.
How to attach an indexed file
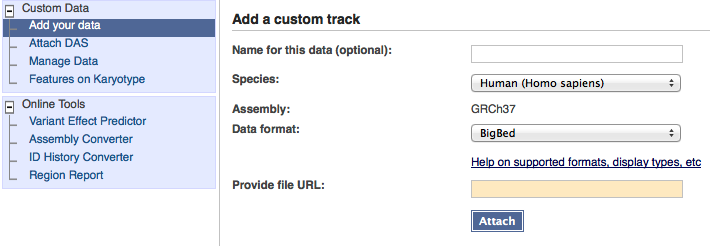
When you select one of these formats, the form button will change to read 'Attach'.
File formats
BAM and CRAM
BAM is a compressed version of the SAM (Sequence Alignment/Map) binary format. BAM uses an index file to give fast access to small sections of the file. CRAM is an improvement on BAM, offering significantly better lossless compression whilst maintaining backwards compatibility with BAM.
Both formats use the new HTSLIB codebase - more information is available from the SAMtools project on github.
bigBed
bigBed is an indexed format created from standard BED files using the bedToBigBed utility program. It allows large datasets to be processed much faster than a conventional BED file.
bigInteract
The bigInteract format is a specialised version of bigBed that uses a standard AutoSQL structure to represent long-range interactions. Please see the UCSC specifications for details of the file structure. In Ensembl, this format is displayed in the same style as the WashU pairwise interaction format.
bigPsl
The bigPsl format is a specialised version of bigBed that uses a standard AutoSQL structure to represent alignments between sequences. Please see the UCSC specifications for details of the file structure. In Ensembl, this format is displayed in the same style as a standard PSL file.
bigWig
The bigWig format is designed for dense, continuous data that is intended to be displayed as a graph. Files can be created from WIG or BedGraph files using the appropriate utility program.
Ensembl currently allows the following configuration options to be set when you upload the file:
- Track colour (10 choices)
- Y-axis maximum and minimum values
VCF format
The VCF format is a tab delimited format for storing variant calls and and individual genotypes. It is able to store all variant calls from single nucleotide variants to large scale insertions and deletions.
More information on this format, which is still under development, can be obtained from the 1000 Genomes Wiki.
Please note that if you are attaching a large VCF file from a URL, you will need to have an index file with the extension .vcf.gz.tbi in the same directory as your data file (and with the same name)
In order to produce the indexed vcf file with the .gz.tbi extension you must follow the following steps:

