Alignments (Text)
Background
Whole genome aligments include "pairwise" sequence alignments between two species, and multi-species alignments using genomes of more than two species.
Export alignments with the "export data" link in the left hand navigation column of the "Genomic alignments" page.
Sequence Display
Only one species is shown by default. Click on select an alignment at the top of the sequence in order to choose an alignment to view.
Chromosomes and scaffolds in the alignment are listed for each species. Sequence is shown under these coordinates. Red, highlighted nucleotides are located in exons.
Click on configure this page at the left of the view to add or change the display. Customisable options are as follows:
- Flanking sequence View the sequence upstream and downstream of the gene.
- Number of base pairs per row Define how many base pairs shown per line.
- Exons Highlight exons from Ensembl, Vega, or ab initio analysis.
- Exons on strand Highlight exons on the forward, reverse or both strands of the chromosome.
- Show variants Show all variants.
- Line numbering Select line numbering. Relative to this sequence starts from 1 for the region displayed, and relative to the coordinate system shows the base pair position using genomic coordinates.
- Conservation regions Highlight regions in which more than 75% of bases match in the alignment.
- Codons Display codons for start and stop translation.
- Display pop-up information on mouseover
Note: Ancestral sequences may be turned off using configure this page.
If the sequence is not known, nucleotides are replaced by dots.
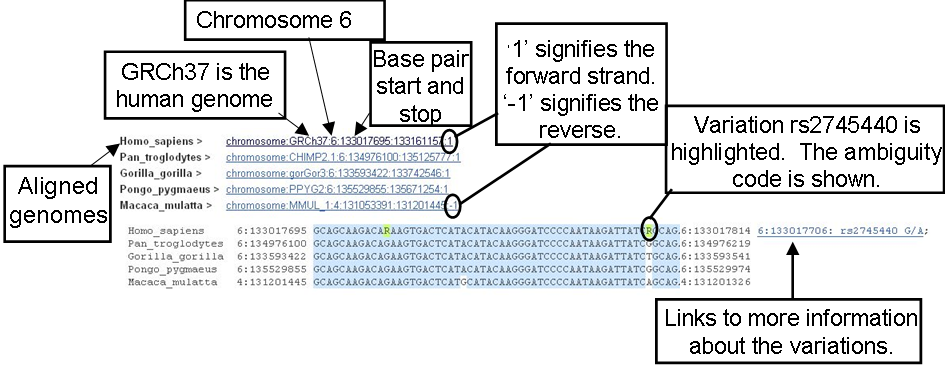
The image above shows the multiple alignments for five catarrhini primates. Conserved nucleotides have been turned on, and are shown by blue highlighting. Variants and line numbering (relative to the coordinate system) are also selected by clicking on the configure this page button.

